Z stacks of FLIM images are usually recorded only for a limited number of Z planes and a limited number of pixels in X and Y. Recording with higher spatial resolution is normally precluded by photo-induced changes in the fluorescence parameters or even by photodamage in the sample. A detailed view at the sample exposure shows, however, that this is not necessarily correct for Z stacks of larger objects, such as small organisms or tissue samples. Here, we show that high-resolution Z stack imaging of an object filling the field of view of an x20 microscope lens is well within the reach of a TCSPC FLIM system. We demonstrate Z stack imaging of a fly (Musca domestica) with 289 Z planes of 2.5 µm Z step width, 1024 x 1024 pixels in X and Y, and 1024 channels in time by using existing functions of the Becker & Hickl DCS-120 confocal FLIM system. Double-exponential decay analysis was performed by the batch processing function of Becker & Hickl SPCImage NG; 3D reconstructions on the basis of the SPCImage results were performed by ImageJ / FIJI.
High Resolution Z-Stack FLIM with the Becker & Hickl DCS‑120 Confocal FLIM System
Wolfgang Becker, Julius Heitz, Lukas Braun, Axel Bergmann, Becker & Hickl GmbH
Abstract: Z stacks of FLIM images are usually recorded only for a limited number of Z planes and a limited number of pixels in X and Y. Recording with higher spatial resolution is precluded by photo-induced changes in the fluorescence parameters or even by photodamage in the sample. A detailed view at the sample exposure shows, however, that this is not necessarily correct for Z stacks of larger objects, such as small organisms or tissue samples. Here, we show that high-resolution Z stack imaging of an object filling the field of view of an x20 microscope lens is well within the reach of a TCSPC FLIM system. We demonstrate Z stack imaging of a fly (Musca domestica) with 289 Z planes of 2.5 µm Z step width, 1024 x 1024 pixels in X and Y, and 1024 channels in time by using existing functions of the Becker & Hickl DCS-120 confocal FLIM system. Double-exponential decay analysis was performed by the batch processing function of Becker & Hickl SPCImage NG. 3D reconstructions on the basis of the SPCImage results were performed by ImageJ / FIJI.
Motivation
The recording of Z stacks of images, or Z scanning has been early introduced into confocal and multiphoton laser scanning microscopes [7]. The microscope scans one image plane, saves the image data, changes the depth of the focus, and scans another plane. The process is continued until the desired number of planes has been scanned, see Fig. 1. Ideally, a sufficiently large number of closely spaced z planes would be scanned to allow the three-dimensional structure of the sample to be reconstructed.

Fig. 1: Principle of Z stack recording
Although Z stack recording is a common procedure in laser scanning microscopy its use in combination with FLIM has not attracted much attention yet. One reason is that FLIM normally focuses on molecular imaging and less on the reconstruction of the spatial structure of a sample. Molecular imaging has to detect small changes in the fluorescence lifetimes, usually in combination with double- and triple exponential decay analysis [1]. The primary information is often in the amplitudes or in the lifetimes of the decay components, not in the apparent lifetime of the decay function. Deriving such information from the decay profiles in the individual pixels requires a large number of photons [4, 6]. Recording these photons not only takes time but also sets tough requirements to the photostability of the sample. Moderate photobleaching may still by acceptable for purely spatial imaging. In FLIM, however, photobleaching induces lifetime changes which make it impossible to derive molecular information from the data. So far, FLIM Z stacks therefore have been recorded with a moderate number of planes only [2, 3]. The data are used to seek out the most promising plane for further decay analysis.
So, can FLIM Z stacks be recorded with a number of planes sufficient to reconstruct the 3D structure of the sample? The options are certainly bleak for 3D imaging of single cells at diffraction-limited X-Y-Z resolution. The energy concentration within the small volume is too high for the cell to survive the procedure. However, if the object of interest is larger, the options are far better. Consider a cell with a diameter and a thickness of 10 µm. The volume is 10-6 mm3. Compare this with the volume of an object completely filling the field of view of an x20 microscope lens. The diameter of the field is about 1 mm. A Z stack over the image area and a depth range of 0.1mm covers an imaging volume of 0.1 mm3. This is 100,000 times the volume of the cell! With the energy spread over the larger volume the average energy density is substantially lower, and so are photobleaching and photodamage effects. High resolution Z stacks of medium-size objects should therefore be well within the reach of a good FLIM system.
FLIM System
To test this hypothesis we used a Becker & Hickl DCS-120 (bh) confocal FLIM system [2, 3]. The DCS-120 has two ps diode lasers of 405 nm and 488 nm, a dual-channel confocal scan head with selectable filters and pinhole sizes attached to a Zeiss Axio Observer microscope, two HPM-100-40 hybrid detectors, and dual-channel SPC-180NX TCSPC / FLIM recording electronics. The lasers, the data acquisition system, and the microscope are controlled by bh SPCM data acquisition software, data analysis is performed by bh SPCImage NG software [6].
The DCS-120 system has two modes for Z stack recording [1, 2]. In both cases the SPCM data acquisition software interacts with the Axio Observer microscope. The FLIM system records a FLIM data set in the current focal plane with a defined acquisition time, saves the data, and then commands the microscope to proceed to the next Z plane. The difference between the modes is in the way the data are saved. The data of each plane can either be written into one element of a data 'Mosaic', or saved into a normal (.sdt) data file of SPCM. Mosaic FLIM has the advantage that no time is lost for saving data, and the data of all planes can be analysed in a single analysis run [1, 6]. However, since the size of the mosaic is limited by memory space, the number of Z planes is limited. Typical z stack formats in this mode are 16 planes (with 512 x 512 pixels x 1024 time channels) to 64 (256 x 256 pixels x 256 time channels). An infinite number of planes can be recorded by saving the planes into individual data files [1]. A disadvantage of this 'Record and Save' procedure can be that saving the data takes time.
Experiment
To be independent of memory limitations we used the record-and-save procedure for our experiment. The essential system setup parameters are shown in Fig. 2. The recording mode is 'FIFO Imaging', with the 'Lifetime+Intensity' option, see Fig. 2 left and second left. 'Lifetime+Intensity' means the number of photons per pixel is determined by a parallel fast counter, thus avoiding nonlinear intensity scale at high count rates [1, 5]. 289 measurement cycles are performed, and each cycle is saved by the 'Autosave' function. The file names are 'Musca‑c0001' through 'Musca‑c0289' (Fig. 2 left). The control parameters for the microscope are shown in Fig. 2, second right. With these settings, the DCS‑120 system records 289 planes from 6047 µm to 6767 µm on the absolute Z scale of the microscope. The Z step with is 2.5 µm. The scan format for the individual planes is 1024 x 1024 pixels, 1024 time channels (Fig. 2, right).
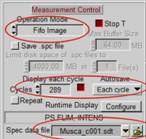
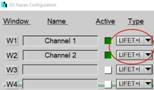
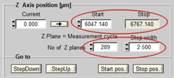
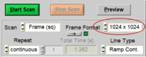
Fig. 2: Essential system parameters of the FLIM system
The measurement object was a fly (Musca domestica, collected from the window sill, no ethics approval required). The excitation wavelength was 405 nm, the excitation power in the sample plane was about 100 µW. The acquisition time per plane was 60 seconds. A 450 nm long-pass filter was used in the detection beam path. The microscope lens was a Zeiss EC PLAN-NEO Fluar M=20, NA=0.5.
Results
Fig. 3 shows images from three planes selected from the 289 planes recorded. The images were selected by the 'Multi-File' view of SPCM. Colour represents lifetime, calculated by the online-lifetime display function of SPCM [1].
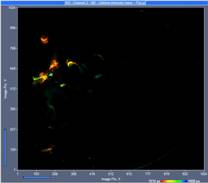
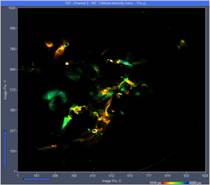
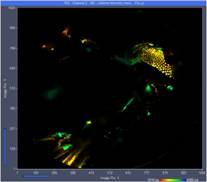
Fig. 3: Three images from different Z planes. Left to right: Plane 120, plane 150, plane 180. Online-lifetime function of SPCM.
At first glance, these images may not look very impressive. The reason is that each of the image represents a thin horizontal slice through the sample only. However, the fact that there are no out-of-plane details visible shows that the DCS-120 scanner optics provides near-perfect suppression of out-of-focus fluorescence.
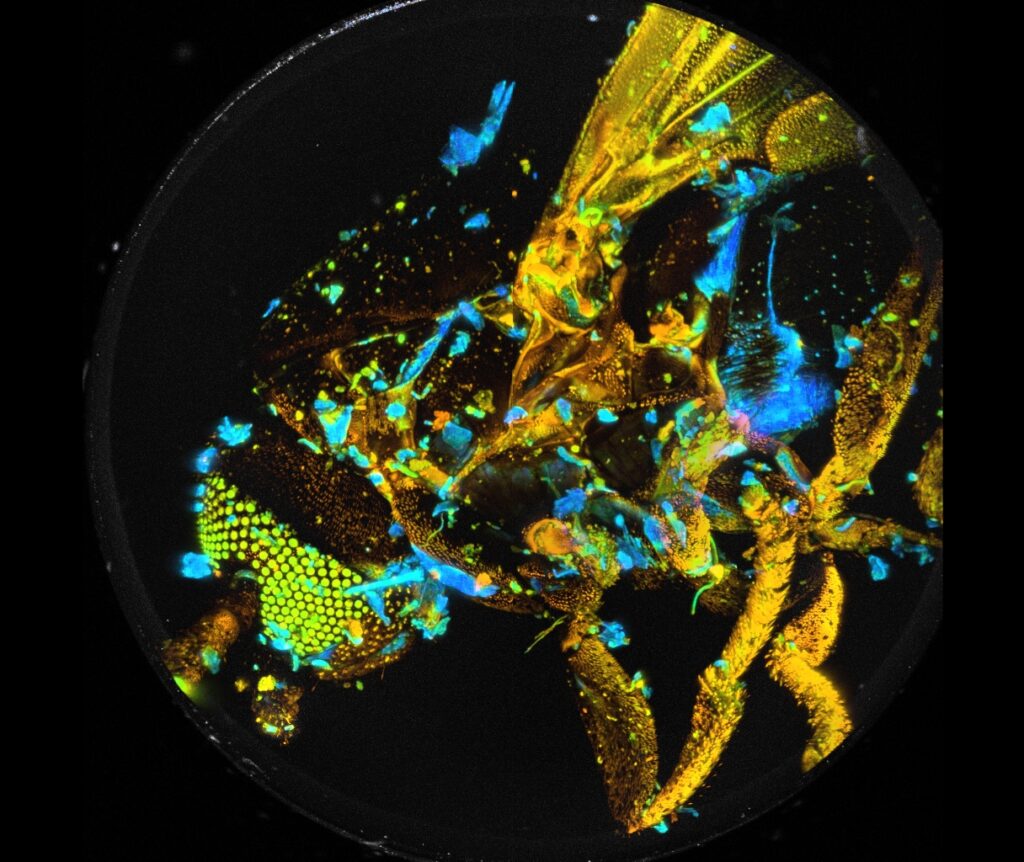
Vertical projections of the data can be created by adding the FLIM data of selected planes. A projection of the FLIM data from all 289 planes is shown in Fig. 4. It gives an impression of the enormous amount of detail contained in the Z stack. Similar projections can be created for selectable ranges of planes. Fig. 5 shows projections for planes 130 to 160 and planes 160 to 194. Since the projections are produced by adding FLIM data of the planes the results can be used like normal FLIM data from a single recording. In particular, they can be sent to or imported into SPCImage NG and processed by multi-exponential decay analysis. This way, amplitude-weighted lifetimes, amplitudes and lifetimes of decay components or ratios of decay parameters can be obtained and displayed as colour-coded images. An example is shown in Fig. 6.
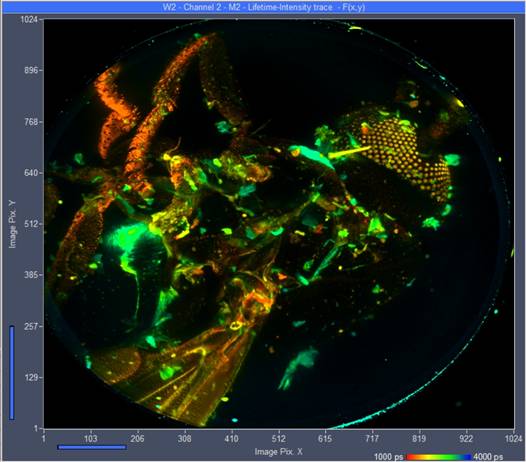
Fig. 4: Vertical projection of all 289 planes into a single FLIM image. Planes added by Multi-File View of SPCM, image displayed by Online-Lifetime function of SPCM. Single-exponential lifetime by first-moment analysis.
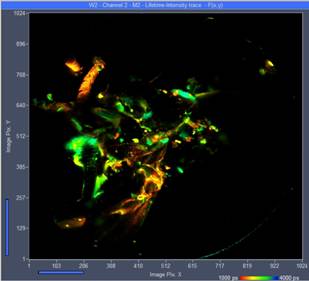
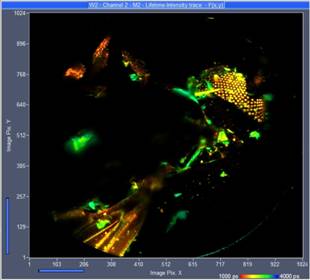
Fig. 5: Vertical projection of planes 130 to 160 (left) and 160 to 194 (right). Single-exponential lifetime by first-moment analysis. Lifetime range from 1000 ps (red) to 4000 ps (blue).
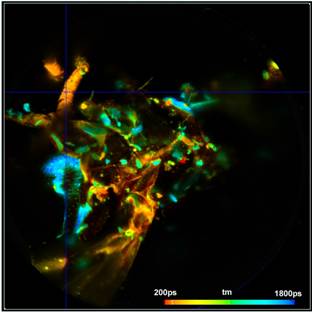
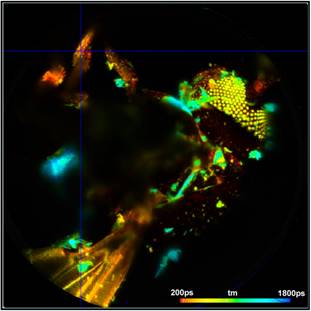
Fig. 6: Vertical projection of planes 130 to 160 (left) and 160 to 194 (right). Data analysis with SPCImage NG. Fit with double-exponential model, amplitude-weighted lifetime.
3D Reconstruction with Image J
3D reconstructions of spatial structures can be obtained by the Image Stacks function of ImageJ in combination with the Batch Processing and Batch Export functions of SPCImage NG. The procedure is as follows. The .sdt files of all planes are loaded into SPCImage NG data analysis software and processed by the 'Batch-Processing' function [6]. The function processes all files with a model function and with model parameters selected before. We recommend to perform a test run on one of the sdt files first to determine the correct IRF, the best model function and an appropriate lifetime and intensity range. We also recommend to run the lifetime analysis on a GPU (Graphics Processing Unit) [6]. Processing a single file then takes only a few seconds, thus keeping the total processing time within reasonable limits.
Once started, batch processing does not require any user interaction. The results are written into subsequent .img files of SPCImage NG. These files are then exported into bitmap files by the 'Batch Export' function of SPCImage NG [6]. Finally, the bitmap files are loaded into ImageJ, and combined by ImageJ's Image Stacks 3D Project routine. The result is a number of 3D presentations from selectable viewing angles.
Two examples are shown in Fig. 7. The data were analysed with a double-exponential model in combination with the 'Incomplete Decay' option of SPCImage. The colour represents the amplitude-weighted lifetime, tm, of the double-exponential decay. The lifetime range is 0 to 1250 ps.
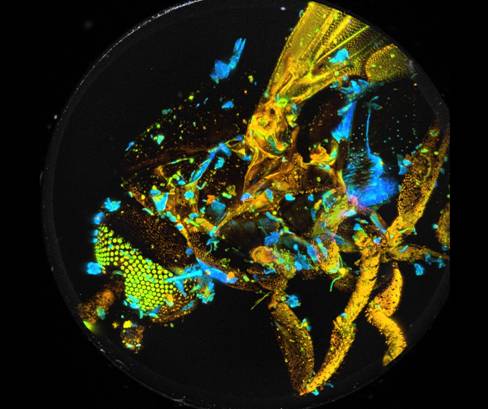
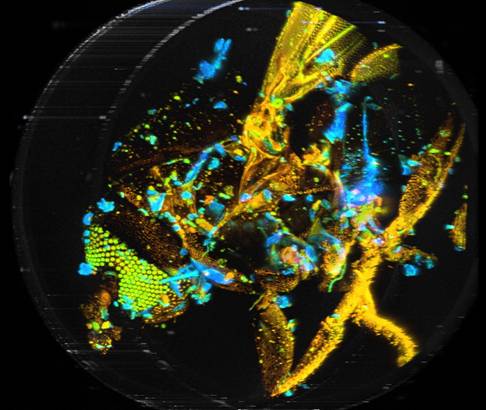
Fig. 7: 3D reconstructions from two different viewing angles. SPCImage NG and ImageJ. Colour represents mean lifetime, tm, of double-exponential decay, lifetime range red to blue = 0 to 1250 ps.
Summary
Photobleaching and photodamage usually preclude the recording of high-resolution FLIM Z stacks of single cells or other objects of micrometer size. However, the requirements to the photostability are significantly relaxed for larger objects. Our results show that a bh TCSPC FLIM system is able to record high-resolution Z stacks of objects with sizes of the field of view of an x20 microscope lens. By using existing functions of the Becker & Hickl DCS-120 confocal FLIM system, we recorded a Z stack of a fly (Musca domestica) with 289 Z planes of 2.5 µm Z step width, 1024 x 1024 pixels in X and Y, and 1024 channels in time. The data of all planes or of a selected range of planes can be projected into a single FLIM image directly in SPCM, and the result be processed by SPCImage NG data analysis. Lifetime analysis of all individual planes can be performed by the Batch Processing function of SPCImage. The results can be further processed by the Image Stacks function of ImageJ / FIJI, providing full 3D reconstructions of the spatio-temporal properties of the sample.
References
1. W. Becker, The bh TCSPC handbook. 10th edition (2023), available on www.becker-hickl.com
3. DCS-120 Confocal and Multiphoton Scanning FLIM Systems, Overview brochure, available on www.becker-hickl.com,
5. Wolfgang Becker, Axel Bergmann, Markus Schubert, Stefan Smietana, Lifetime-intensity mode delivers better FLIM images. Application note, available on www.becker-hickl.com.
6. SPCImage NG data analysis software. In: W. Becker, The bh TCSPC handbook. 10th edition (2023)
7. J. Pawley (ed.), Handbook of biological confocal microscopy, 3rd edn., Springer (2006)
Contact:
Wolfgang Becker
Becker & Hickl GmbH
Berlin, Germany
Nunsdorfer Ring 6-9
Email: becker@becker-hickl.com